MolBoolean: Beyond Proximity Ligation
MolBoolean™ is a novel in situ proximity technology developed by Atlas Antibodies that enables the simultaneous detection of both free and interacting fractions for two protein targets.
MolBoolean™, an in situ proximity technology by Atlas Antibodies, introduces a new approach to protein interaction analysis, offering an alternative to proximity ligation assay (PLA) and expanding research possibilities.
Understanding protein interactions is fundamental to unraveling cellular mechanisms in health and disease. Traditional methods, such as proximity ligation assay (PLA), have been instrumental in detecting protein-protein interactions, but they come with limitations in complexity and throughput.
To address these challenges, Atlas Antibodies, in collaboration with Professor Ola Söderberg, has introduced MolBoolean™, a novel in situ proximity technology that revolutionizes protein interaction analysis by providing a more straightforward, scalable, and robust alternative.
The MolBoolean™ assay applies a Boolean logic at a molecular level to distinguish between free and interacting proteins, mapping protein interactions with high spatial specificity. The OR logic identifies either protein A or B independently, signaling that they are present but not necessarily interacting. The AND logic is applied when proteins A and B are interacting; they are detected together as complex AB, indicating a combined “true” interaction.
MolBoolean™ represents a novel approach that circumvents key limitations of PLA by allowing for the detection and quantification of protein interactions in a binary manner. Unlike traditional methods MolBoolean™ enables the simultaneous detection and spatial quantitative analysis of both free and interacting fractions for two protein targets in cells and tissue (~40 nm proximity).
In contrast, traditional Proximity Ligation Assay (PLA) methods primarily detect interacting proteins without providing information on the free, non-interacting fractions. PLA also lacks the built-in data normalization to total protein levels, which can lead to less accurate quantification, especially when protein expression levels vary. Additionally, PLA may require engineered protein expression and often involves more complex workflows, potentially limiting its applicability in certain experimental setups.
By addressing these limitations, MolBoolean™ provides a more comprehensive and accurate analysis of protein-protein interactions, enhancing the depth and reliability of data in biomedical research.
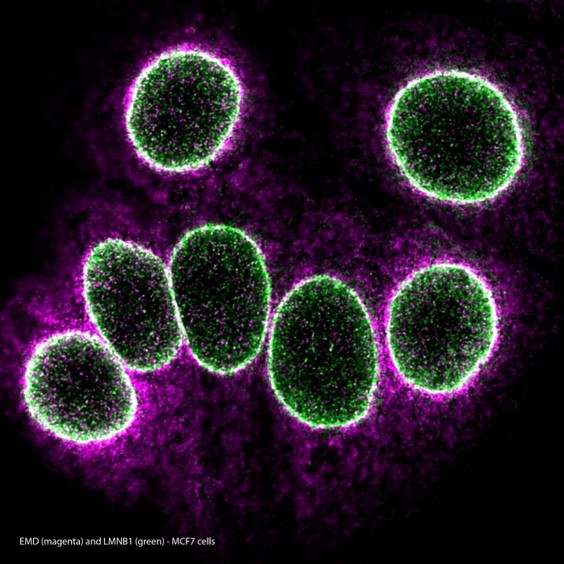
Left: EMD/LMNB1 MolBoolean staining in MCF7 cells
MolBoolean™ analysis of the interaction (visible in white) between EMD (magenta) and LMNB1 (green) in MCF7 cells, using the monoclonal anti-EMD (Cat. AMAb90562) and the polyclonal anti-LMNB1 (Cat. HPA050524) antibodies from Atlas Antibodies AB. Image shows the spatial location of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels.
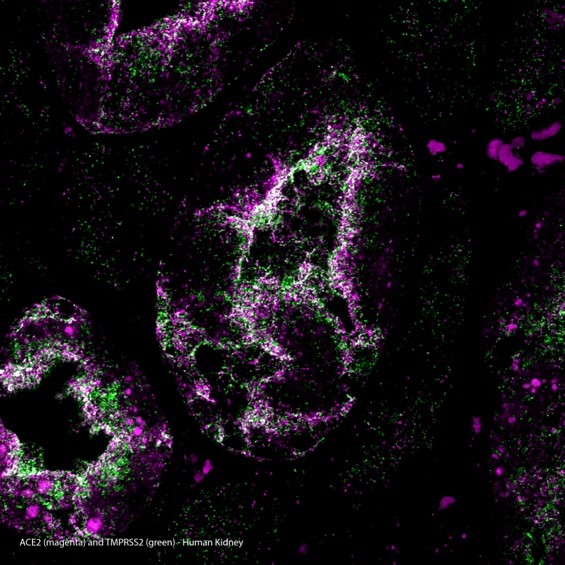
Right: ACE2/TMPRSS2 MolBoolean staining in human kidney
MolBoolean™ analysis of the interaction (visible in white) between ACE2 (magenta) and TMPRSS2 (green) in human kidney, using the monoclonal anti-ACE2 (Cat. AMAb91259) and the polyclonal anti-TMPRSS2 (Cat. HPA035787) antibodies from Atlas Antibodies AB. Image shows the spatial location of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels.
MolBoolean™ offers significant benefits in fields requiring complex spatial interaction and quantification data, such as:
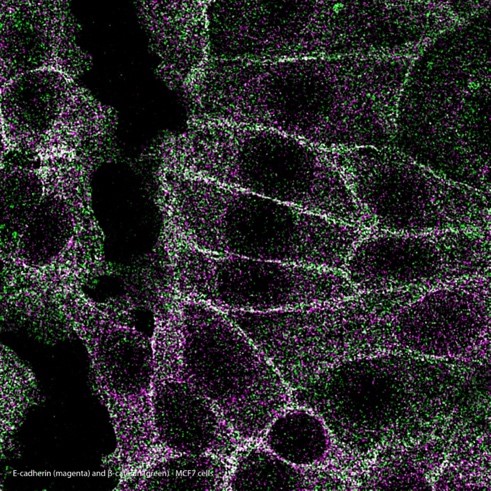
Left: E-cadherin/ β-catenin MolBoolean staining in MCF7 cellsMolBoolean analysis of the interaction (visible in white) between E-cadherin (magenta) and β-catenin (green) in MCF7 cells, using the monoclonal anti-CDH1 (Cat. AMAb90862) and the polyclonal anti-CTNNB1(Cat. HPA029159) antibodies from Atlas Antibodies AB. Image shows the spatial location of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels.
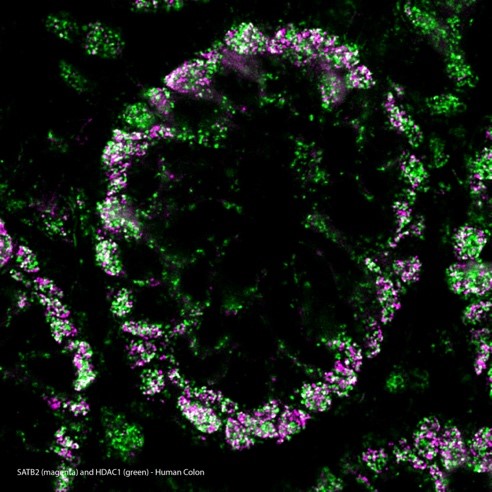
Right: SATB2/HDAC1 MolBoolean staining in human colon
MolBoolean™ analysis of the interaction (visible in white) between SATB2 (magenta) and HDAC1 (green) in human colon, using the monoclonal anti-SATB2 (Cat. AMAb90682) and the polyclonal anti-HDAC1 (Cat. HPA029693) antibodies from Atlas Antibodies AB. Image shows the spatial location of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels.
MolBoolean™ holds immense potential across various biomedical fields, including cancer research, neuroscience, and immunology. Its ability to quantify dynamic protein interactions makes it particularly valuable for:
One of the core advantages of MolBoolean™ is its ability to work efficiently with standard immunocytochemistry and flow cytometry platforms, making it accessible to a wider range of researchers. This method streamlines the process of interaction detection while preserving the spatial information critical to cellular studies.
Rivas-Santisteban R, et al. (2024): MolBoolean was used to analyze the impact of GPR88 on kappa opioid receptor signaling in a heterologous system and primary striatal neurons. The results revealed that GPR88 significantly impaired kappa opioid receptor function, suggesting a novel regulatory mechanism in opioid signaling.
Kotliar IB, et al. (2024): This study employed MolBoolean to perform multiplexed mapping of G-protein-coupled receptors (GPCRs) interacting with receptor activity-modifying proteins (RAMPs). The technology enabled precise quantification of these interactions, uncovering new insights into GPCR modulation.
Malmqvist M, et al. (2024): MolBoolean was applied to quantify interactions between dopamine D2 and adenosine A2A receptors in rat brain. The findings demonstrated a significant proportion of these receptors forming complexes, offering critical insights into their role in neurological disorders.
Raykova D, et al. (2023): This study introduced and validated the MolBoolean method for Boolean analysis of protein interactions at a molecular level. The results confirmed its high specificity and reliability, establishing it as a robust alternative to existing interaction detection techniques.
Rivas-Santisteban R, et al. (2023): Using MolBoolean, researchers demonstrated a high proportion of dopamine D2 receptors interacting with adenosine A2A receptors in striatal medium spiny neurons of Parkinson’s disease models. This finding supports the therapeutic potential of targeting these receptor interactions in neurodegenerative diseases.
As research in these areas continues to expand, MolBoolean™ is expected to become an essential tool for advancing discoveries in molecular biology and precision medicine.
The introduction of MolBoolean™ by Atlas Antibodies marks a significant advancement in the study of protein interactions, providing researchers with a powerful, efficient, and scalable alternative to PLA. By simplifying workflows and improving data reliability, this technology paves the way for more comprehensive and high-throughput studies in biomedical research.
Rivas-Santisteban R, et al, (2024) GPR88 impairs the signaling of kappa opioid receptors in a heterologous system and in primary striatal neurons. Neuropharmacology. 2024 Nov 27:110242. doi: 10.1016/j.neuropharm.2024.110242. Epub ahead of print. PMID: 39613254.
Kotliar IB, et al, (2024) Multiplexed mapping of the interactome of GPCRs with receptor activity-modifying proteins. Sci Adv. 2024 Aug 2;10(31):eado9959. Epub 2024 Jul 31. PMID: 39083597; PMCID: PMC11290489.
Malmqvist M, et al, (2024) Quantifying dopamine D2 and adenosine A2A receptor interactions in rat brain using MolBoolean™ technology. #8217 Society for Neuroscience, SfN, (2024).
Raykova D, et al, (2023) A method for Boolean analysis of protein interactions at a molecular level. Nat Commun. 2023 Sep 6;14(1):5450. doi: 10.1038/s41467-023-41325-3. Erratum for: Nat Commun. 2022 Aug 13;13(1):4755. doi: 10.1038/s41467-022-32395-w. PMID: 37673885; PMCID: PMC10482831. Rivas-Santisteban R, et al, (2023) Boolean analysis shows a high proportion of dopamine D2 receptors interacting with adenosine A2A receptors in striatal medium spiny neurons of mouse and non-human primate models of Parkinson’s disease. Neurobiol Dis. 2023 Nov;188:106341.doi: 10.1016/j.nbd.2023.106341. Epub 2023 Oct 31. PMID: 37918757.
We gladly support you by keeping you updated on our latest products and the developments around our services.