What are the differences between antibody types?
Discover the differences between polyclonal, monoclonal and recombinant antibodies
MolBoolean™ is a novel in situ proximity technology developed by Atlas Antibodies that enables the simultaneous detection of both free and interacting fractions for two protein targets.
The dynamic mechanisms of protein-protein interactions (PPIs) in cells and tissue are heavily dependent on the spatial location, but also the abundance of the respective interacting partners. Therefore, for in situ protein proximity studies, it is pivotal to not only understand the number of interaction events, but also the number of ingoing components.
Working closely together with Professor Ola Söderberg at Uppsala University, Atlas Antibodies has developed the MolBoolean™ assay, a novel in situ protein proximity technology for simultaneous detection of both interacting and non-interacting
fractions of two protein targets in cells and tissue,based on the ‘Boolean logic’: OR, AND, NOT (Figure 1).
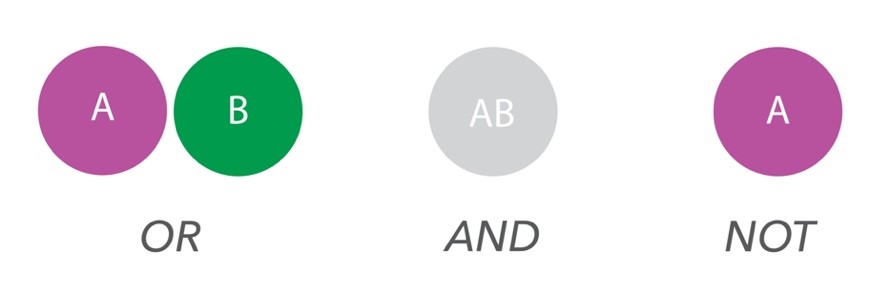
MolBoolean™ relies on dual target recognition with proximity probes and generates rolling circle amplification products to increase signals, meaning that even low abundant proteins can be detected.
On top of that, by performing a series of proprietary molecular steps, MolBoolean™ differentiates between amplified products stemming from individual versus interacting proteins, and this is indicated by the binding of one or two separate fluorescent detection reporters. The more comprehensive extraction of information provided by MolBoolean™ enables normalization of the interaction data to total target protein levels.
MolBoolean™ offers several advantages over traditional methods like in situ PLA, addressing key gaps in data interpretation and providing more reliable results:
• Normalization of Data
With MolBoolean™, the number of protein interactions can be normalized to the total number of target proteins. This is crucial because target protein levels can be influenced by various factors such as cell treatments or disease states. By providing a normalized measure, MolBoolean™ allows for more accurate comparisons between samples.
• Detection of Interacting and Non-interacting Fractions
Unlike traditional methods, MolBoolean™ can detect both interacting and non-interacting endogenous fractions for two protein targets in cells and tissues. The more comprehensive detection capability increases the spatial information that can be extracted from the experiment, allowing researcher to determine locations of positive or negative protein interactions.
• Consistent Molecular Process:
The Rolling Circle Products (RCPs) stemming from free or interacting proteins are generated through the same molecular process steps. This ensures uniform signal efficiencies, reducing uncertainty during data analysis.
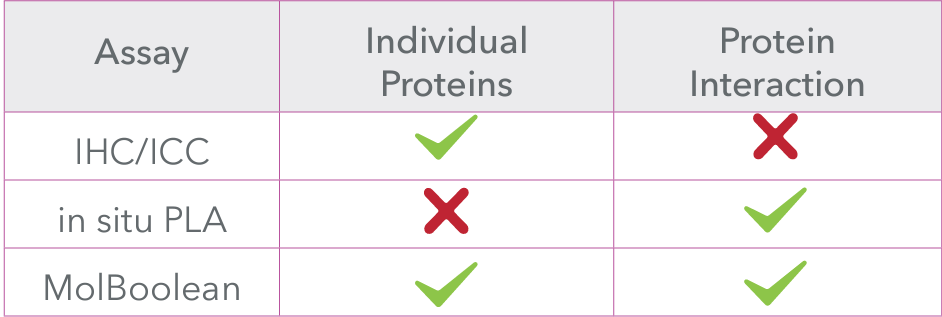
Overall, MolBoolean™ offers a more reliable and comprehensive solution for detecting protein interactions, addressing key gaps in data interpretation, and providing researchers with more accurate insights into molecular interactions in cells and tissues (Figure 2).
MolBoolean’s versatility and precision in studying protein-protein interactions offer a multifaceted approach to therapeutic innovation.
From targeted drug development to personalized medicine and biomarker discovery, MolBoolean™ holds immense potential to advance biomedical research and translate scientific findings into tangible clinical benefits for patients.
We gladly support you by keeping you updated on our latest products and the developments around our services.